



API Suppliers
0

US DMFs Filed
0

CEP/COS Certifications
0

JDMFs Filed
0
Other Certificates
0
Other Suppliers
0
0

USA (Orange Book)
0

Europe
0

Canada
0

Australia
0

South Africa
0
Uploaded Dossiers
0
U.S. Medicaid
0
Annual Reports
0
0
USFDA Orange Book Patents
0
USFDA Exclusivities
0
Blog #PharmaFlow
0
News
0
EDQM
0
USP
0
JP
0
Other Listed Suppliers
0
0
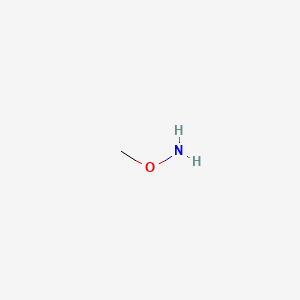

1. Methoxyamine
2. Methoxyamine Hydrochloride
3. Methoxylamine
1. Methoxyamine
2. 67-62-9
3. Methoxylamine
4. Hydroxylamine, O-methyl-
5. Hydroxylamine Methyl Ether
6. Nci-c60060
7. Ch3onh2
8. 9tzh4wy30j
9. Alpha-methylhydroxylamine
10. Hsdb 2883
11. Einecs 200-660-1
12. Unii-9tzh4wy30j
13. Brn 1098249
14. Methoxy Amine
15. N-methoxyamine
16. Meonh2
17. Nh2ome
18. O-methyl Hydroxylamine
19. O-methyl-hydroxylamine
20. O-methylhydroxyl Amine
21. Spectrum_001650
22. O-methyl Hydroxyl Amine
23. Specplus_000744
24. Spectrum2_000914
25. Spectrum3_000765
26. Spectrum4_001103
27. Spectrum5_001275
28. Methoxyamine [mi]
29. Bspbio_002410
30. Kbiogr_001546
31. Kbioss_002130
32. 4-01-00-01252 (beilstein Handbook Reference)
33. Divk1c_000600
34. Divk1c_006840
35. Methoxyamine [who-dd]
36. Spbio_000788
37. Chembl1213633
38. Dtxsid8043862
39. Gmpkipwjbdourn-uhfffaoysa-
40. Kbio1_000600
41. Kbio1_001784
42. Kbio2_002130
43. Kbio2_004698
44. Kbio2_007266
45. Kbio3_001630
46. Chebi:192842
47. Ninds_000600
48. O-methylhydroxylamine [hsdb]
49. Bbl028003
50. Mfcd00044753
51. Stl373472
52. Akos009087974
53. Db06328
54. Idi1_000600
55. Ncgc00178666-01
56. Bp-31110
57. F16476
58. Q18344129
| Molecular Weight | 47.057 g/mol |
|---|---|
| Molecular Formula | CH5NO |
| XLogP3 | -0.7 |
| Hydrogen Bond Donor Count | 1 |
| Hydrogen Bond Acceptor Count | 2 |
| Rotatable Bond Count | 0 |
| Exact Mass | 47.037113783 g/mol |
| Monoisotopic Mass | 47.037113783 g/mol |
| Topological Polar Surface Area | 35.2 Ų |
| Heavy Atom Count | 3 |
| Formal Charge | 0 |
| Complexity | 4.8 |
| Isotope Atom Count | 0 |
| Defined Atom Stereocenter Count | 0 |
| Undefined Atom Stereocenter Count | 0 |
| Defined Bond Stereocenter Count | 0 |
| Undefined Bond Stereocenter Count | 0 |
| Covalently Bonded Unit Count | 1 |
Investigated for use/treatment in cancer/tumors (unspecified).
Methoyxamine is investigated for use as an adjunct to alkylating agents, reverse resistance to chemotherapy, and enhancing radiation therapy. Methoxyamines proposed mechanism of action is through blocking of the abasic sites (apurinic/apyrimidinic - AP sites) created by the cleavage of base excision repair (BER) glycoslyates. DNA alkylating agents cause cell death through excessive DNA damage by adduct formation. The human mechanism for DNA repair is very efficient and cancer therapeutics which use this mechanism are often ineffective due to resistance by efficient repair mechanisms such as base excision repair (BER). Alkylating agents such as tezmozolomide form methylated DNA adducts such as O6-methylguanine (O6mG), 7-methylguanine (N7mG) and 3-methyladenine (N3mA). O6mG is a cytotoxic and genotoxic adduct which is repaired by O6-methylguanine DNA-methyltransferase (MGMT). O6mGs cytotoxicity is due to the mismatch repair mechanism (MMR), but cell induced defects in this repair pathway can lead to drug resistance. The N7mG (dominant lesions caused by methylating agents) and N3mA adducts are both repaired by the BER mechanism. Methoxyamine disrupts the BER pathway, increasing the amount of cytotoxic adducts, which results in cell death. Methoxyamine inhibits BER by stabilizing the AP sites created by cleavage of BER glysosylates, forming MX-AP lesions. Methoxyamine may be an effective adjunct to iododeoxyuridine(IUdR) induced radiosensitization and radiation treatment. IUdR is a halogenated pyrimidine which is incorporated into cellular DNA instead of thymidine, which enhances radiotumor sensitivity. Methoxyamine is proposed to have a dual action in this treatment as it alters cell cycle kinetics as well as prevents repair of DNA by BER, allowing increased sensitivity of tumor cells to DNA damage by radiation therapy. The efficiency of cell cycle repair has been shown to be cell cycle dependent, with the G1 phase being second most sensitive to ionizing radiation (the mitotic, M, phase is the most sensitive). Methoxyamine increases the amount of protein 53 (P53) and protein Rb (pRB), senescence factors which cause the cell to remain in the G1 phase. Methoxyamine also creates a stringent checkpoint at the G1/S boundary as well as an insufficient checkpoint at the G2 stage, preventing cells from going into the S phase. The increased number of G1 cells makes methoxyamine treated tumors more susceptible to ionizing radiation. The temozolomide and methoxyamine created lesion MX-AP not only disrupts the BER pathway but inhibits topoisomerase II alpha (topo II), an enzyme necessary for DNA replication, recombination and chromosome segregation. MX-AP sites block DNA replication and interfere with choromosome splitting. It is currently uncertain how what the interaction between topoisomerase II and methoxyamine causes cytotoxicity, but several mechanisms have been proposed, such as MX-AP sites binding to topo II, thus reducing their functionality by forming a toxic complex.
THE MAJOR MUTAGENIC EFFECT OF O-METHYLHYDROXYLAMINE ON VEGETATIVE SD PHAGE IN ESCHERICHIA COLI IS PROBABLY DUE TO REPLICATION ERRORS RESULTING FROM INCORPORATION OF ENZYMICALLY MODIFIED PRECURSORS INTO DNA.
BUDOVSKII EI ET AL; THE MECHANISM OF THE MUTAGENIC ACTION OF HYDROXYLAMINE. XIV. O-METHYLHYDROXYLAMINE-INDUCED DIRECT MUTATIONS IN VEGETATIVE SD PHAGE; MUTAT RES 59(2) 285 (1979)
THE DIPHOSPHATE REACTION PRODUCT (MO6A) OF O-METHYLHYDROXYLAMINE WAS PREPARED AND COPOLYMERIZED WITH CYTIDINE DIPHOSPHATE OR ADENOSINE DIPHOSPHATE. THE SPECIFIC CHANGED BASE PAIRING IS PROBABLY CAUSED BY A TAUTOMERIC SHIFT FROM THE AMINO TO THE IMINO. CHANGING THE TAUTOMERIC EQUILIBRIUM OF A PURINE BY A SUBSTITUTION REACTION WITH A MUTAGEN SHOULD BE CONSIDERED AS A CONTRIBUTING FACTOR FOR THE MECHANISM OF MUTATION OF HYDROXYLAMINES.
SINGER B, SPENGLER S; REACTION OF O-METHYLHYDROXYLAMINE WITH ADENOSINE SHIFTS TAUTOMERIC EQUILIBRIUM TO CAUSE TRANSITIONS; FEBS LETT 139(1) 69 (1982)


